Quantitation: Overview
Many different approaches to protein quantitation using mass spectrometry data have been
described in the literature. For a short, recent review, see
Ong, S. E. and Mann, M., Mass
spectrometry-based proteomics turns quantitative, Nature Chemical Biology 1 252-262 (2005).
In terms of the "mechanics" of their implementation, most of the popular approaches
can be classified into a relatively small number of protocols:
- Reporter: Quantitation based on the relative intensities
of fragment peaks at fixed m/z values within an MS/MS spectrum. For example,
iTRAQ and
Tandem Mass Tags
- Precursor: Quantitation based on
the relative intensities of extracted ion chromatograms (XICs)
for precursors within a single data set. This is by far the most widely used approach, which can be used
with any chemistry that creates a precursor mass shift. For example,
18O,
AQUA,
ICAT,
ICPL,
Metabolic,
SILAC,
etc., etc.
- Multiplex: Quantitation based on the relative
intensities of sequence ion fragment peaks within an MS/MS spectrum. This is a
novel
approach, which can be used with labels located at the peptide terminus,
such as 18O or SILAC at K or R in combination with tryptic digestion.
- Replicate: Label free quantitation
based on the relative intensities of extracted ion
chromatograms (XICs) for precursors in multiple data sets aligned using mass and elution time.
- emPAI: Label free quantitation for the
proteins in a mixture based on protein coverage by the peptide matches in a database
search result.
- Average: Label free quantitation
for the proteins in a mixture based on the
application of a rule to the intensities of extracted ion chromatograms (XICs) for the
peptide matches in a database search result.
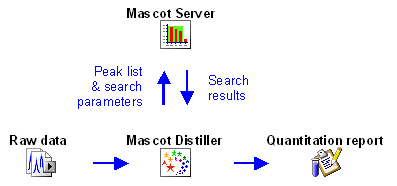
Some protocols can be fully implemented within a
Mascot result report because all the necessary information is present in the peak list.
These protocols are Reporter,
Multiplex, and
emPAI. In fact, emPAI is "always on", and will be
reported whenever an MS/MS search contains at least 100 spectra.
The other three protocols require additional information from the raw data file, either because
it is necessary to integrate the elution profile of each precursor peptide or because
information is required for precursor peptides that were not used to trigger MS/MS scans,
so are missing from the peak list. So, for Precursor,
Replicate, and
Average, the quantitation report is generated in
Mascot Distiller, which has access to both the Mascot search results and the raw data.
Besides the choice of protocol, there are a large number of other choices and
parameters associated with searching, processing, and reporting of quantitation data.
These choices and parameters are necessary to provide sufficient flexibility and control,
yet it would be undesirable to expose all of them in the search form.
The solution is to encapsulate all the settings for a quantitation experiment into a
named quantitation method. This means that
quantitation support requires just a single control in the search form:
The set of quantitation methods is defined by an XML configuration file, called
quantitation.xml.
As with other configuration files, this file lives on the Mascot Server and is downloaded by Mascot
Distiller and other clients as required. Brave souls may choose to edit the XML file directly, but
a more friendly interface is provided by a browser based
configuration editor.
 Mascot takes its modification definitions direct from an XML representation of the
Unimod database. To update the local
definitions, simply download the latest XML file from the Unimod
help page.
Mascot takes its modification definitions direct from an XML representation of the
Unimod database. To update the local
definitions, simply download the latest XML file from the Unimod
help page.
In Unimod, both amino acid residues and modifications are defined in terms of their elemental
composition. This is important for metabolic labelling, in which the isotopic
label is present throughout the peptide backbone.
Unimod also provides a framework for
including local definitions of modifications within a quantitation method. For example,
the multiplex method may require that a modification has two neutral losses. One of 0 Da and
one corresponding to the complete modification moiety, so that a mixed spectrum, containing both labelled
and unlabelled peptides, can be matched with a good score. It would be confusing to have
such an artificial modification appearing in Unimod, so the preference is to define it within
the quantitation method.
In the context of a quantitation method, Mascot now supports exclusive modifications.
A group of
exclusive modifications can be thought of as a choice of fixed modifications.
In many quantitation experiments, separate samples are derivatised then pooled. Thus, a given
peptide may carry one or the other set of modifications, but never a mixture of both. Some people
use the term "binary" for this type of specificity. We prefer exclusive because binary implies
only two possibilities. A SILAC experiment might have three or four different labels which will
never be mixed on a single peptide. One of reasons for introducing this new type of modification
is that variable modifications
greatly increase the size of the search space, because all of the possible permutations
and combinations of modified and unmodified residues have to be explored. As the search space
becomes larger, the search
takes longer and the score threshold increases, making it more difficult to get significant
matches from marginal spectra. By treating labels as a choice of fixed modifications,
we avoid this combinatorial explosion.
Two key concepts have already been introduced. A quantitation method
encapsulates all the settings for searching, processing, and reporting of quantitation data. The
"mechanics" of the method are specified by the protocol.
These keywords represent concepts, and also structures in the configuration file. You will see these
words used consistently, (we hope), in this help, in the browser based configuration editor, and
in the XML configuration file.
Another important concept and keyword is a component.
This is the characteristic property of a peptide that identifies its origin in the sample mixture. For example,
in a SILAC experiment, one component might be unmodified peptides while another component is peptides
modified with Label:13C(6) on arginine or lysine.
If the protocol was reporter, then a component would be identified by a reporter ion m/z value. If
it was a metabolic labelling experiment, then one component might be 14N peptides and the other
15N peptides. In a label free experiment, using the replicate protocol, each data file would be a
component.
If the one component was called light and the second heavy,
we might want a report to list the ratio of heavy over light, or maybe light over heavy.
This is specified as a report ratio
and is not limited to two components. In an 18O experiment, for example, you might want to report
the ratio of (18O1 + 18O2) / 18O0. Both the numerator
and denominator of a reported ratio can be linear combinations of components. For example,
(0.5 * A + 0.5 * B - C) / (D + E) ... not that we can think of a practical use for something so complicated.
In a quantitation method, modifications are organised into groups,
classified as fixed, variable, or exclusive. Modification groups can be defined
as variable or exclusive at the component level, where they usually characterise the component.
They can also be defined at the method level, but only as fixed or variable. Defining modifications
at the method level is a convenience, for modifications that are important to the method, and saves having
to choose them in the search form.
The other method level tabs in the configuration editor, which correspond to child elements of the method element
in the XML file, are:
- integration: Choices and parameters to control the way in which extracted
ion chromatograms are integrated in Mascot Distiller. Also, how to align precursors in the replicate protocol.
- quality: Miscellaneous quality criteria that peptide matches must
meet before they can be used for quantitation. The most important is the strength of the peptide match,
defined in terms of either a minimum score, a maximum expect value, or the score being at or above either
the identity threshold or the homology threshold.
- outliers: When ratios for individual peptide matches are combined into
ratios for protein hits, a variety of procedures are available for detecting and rejecting
outliers
- normalisation: Whatever the quantitation method, it can be difficult
to ensure that each component is treated identically.
If it is reasonable to expect that only a minority of the proteins in the sample will be up or down regulated,
a common solution is to normalise the data so as to make the average ratios across the entire
data set unity.
|