Quantitation: Report format
When the quantitation method is either Reporter or Multiplex, additional formatting controls are displayed near the top of the Mascot
result report. After making changes to any of these controls, choose the Format As button to reload the report using the new settings.
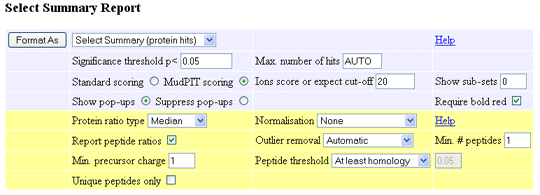
Protein ratio type: A drop down list with the choices Average, Median, and Weighted.
- Average: The protein ratio is the geometric mean of the peptide ratios
- Median: The median peptide ratio is selected to represent the protein ratio. If
there are an even number of peptide ratios, the geometric mean of the median pair is used
- Weighted: For each component, the intensity values of the assigned peptides are summed
and the protein ratio(s) calculated from the summed values. This gives a weighted average,
which will be the best measure if the accuracy is limited by counting statistics. When a
weighted average is reported, the standard deviation is the weighted standard deviation.
Normalisation: A drop down list with the choices None, Average ratio, Median ratio, and Summed intensities.
- None: No normalisation is performed
- Average ratio: For each peptide ratio, a correction factor is applied such that the geometric
mean of the ratios for all peptide matches that pass the quality tests is unity
- Median ratio: For each peptide ratio, a correction factor is applied such that the median of
the ratios for all peptide matches that pass the quality tests is unity
- Summed intensities: (Reporter protocol only) A correction factor is applied such that the sum
of the intensities for a reporter ion peak over all peptide matches that pass the quality tests is the
same for all the reporter ions
Report peptide ratios: When checked, ratios for individual peptides are included in the peptide match tables.
Outlier removal: The available methods for testing and removing outliers are None, Automatic, Dixon's method,
Grubbs' method, and Rosner's method. Choosing auto means that Dixon's method will be used if the number of values is between 4 and 25,
while Rosner's method will be used if the number of values is greater than 25. For a detailed description of each method,
refer to the Statistical procedures page.
Min. # peptides: The minimum number of valid peptide match ratios that are required in order to report a protein
hit ratio.
Min. precursor charge: A peptide ratio will not be reported if the precursor charge is less than this value. Note
that this number is compared with the absolute precursor charge. If you are analysing negative ion data, and the setting is 2, a peptide match
with a charge of [M-3H]3- will be used for quantitation, while a match with a charge of [M-H]- will not.
Peptide threshold: A peptide match will only be used for quantitation if it meets this criterion. The choice is
Minimum Score, Maximum expect, At least identity (threshold), and At least homology (threshold). For the first two choices,
an appropriate value must be entered in the adjacent edit box.
Unique peptides only: When checked, ratios are only reported for peptide sequences that are unique to one protein hit,
(which may contain more than one protein database entry). See the
configuration help page for additional information.
When the quantitation method is either Reporter or Multiplex, there are several additions to the main Mascot result report.
Quantitation related information is presented on a yellow background, in four main areas: the header, the format controls,
(described above), a quantitation summary for each protein hit, and ratios for individual peptides, (optional).
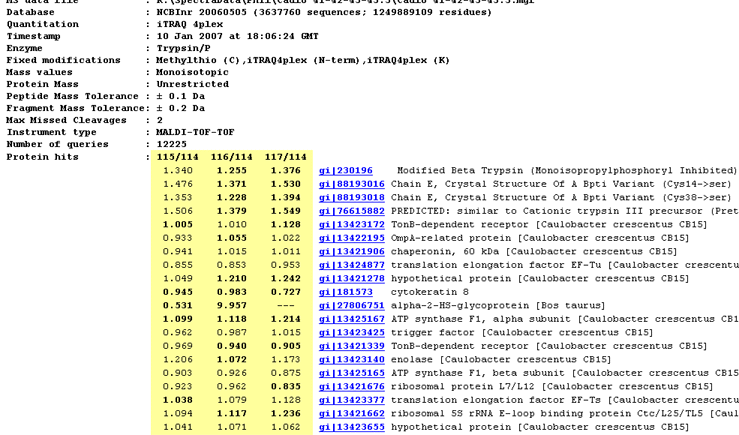
The report header includes a list of the protein hits tabulated in the body of the report. Where quantitation information
is available, it is listed alongside each protein. The ratios to be reported and the column headings are specified as
part of the quantitation method.
- If a ratio is shown in bold face, it is significantly different from 1 at a 95% confidence level.
More details can be found on the Statistical procedures page.
This may or may not be meaningful.
In this screen shot, for example, the top hits are trypsin variants, so the quantitation information is simply
irrelevant.
- If a ratio is missing, this will usually be because the number of valid peptide match ratios was less than the
stipulated minimum.
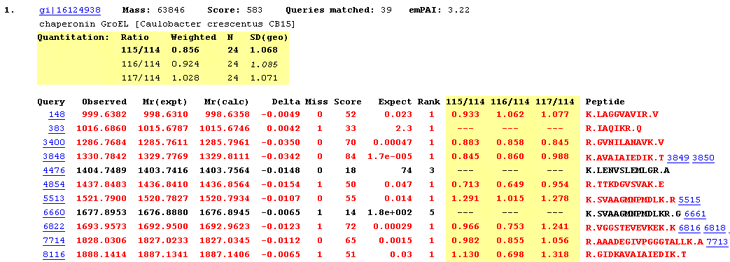
For each protein hit, a table shows the average ratio(s) for the protein, together with the number of peptide
ratios that contributed, N, and the geometric standard deviation, SD(geo). If the number of valid peptide
match ratios was less than the stipulated minimum, dashes will be displayed. If the peptide match ratios do not
appear to be a sample from a
normal distribution, the geometric standard deviation
will be displayed in italics, and will never be bold, because it must be considered unreliable. As mentioned previously,
bold face indicates a ratio that is significantly different from 1 at a 95% confidence level.
Since we are dealing with ratios, the average is the geometric mean
and the standard deviation is the geometric standard deviation, which is a factor.
In other words, the confidence interval is obtained by dividing and multiplying the average by the standard
deviation, which is never less than 1.0. For example, if the average is 1.055 and SD(geo) is
1.028 then the 95% confidence interval is 1.026 to 1.084.
Warning: The standard deviation reported here measures the variance
within the data submitted for a single Mascot search. This will often be tiny compared with the variance between technical
replicates, (i.e. repeated analyses of this same sample). This, in turn, will invariably be tiny compared with
the variance between biological replicates, (analyses of samples from different subjects or different treatment
groups). It can be dangerous to read too much into small changes within a single search when the biological variance is huge.
For a more detailed discussion, see
Karp, N. A., et al., Impact of replicate types
on proteomic expression analysis, Journal of Proteome Research 4 1867-1871 (2005).
Individual peptide match ratios will be displayed if the Report peptide ratios checkbox is checked. Dashes
are displayed when a ratio cannot be determined. This may be because one or more of the relevant peaks were missing,
giving a ratio which was zero, infinity, or indeterminate. Alternatively, the peptide match may have been rejected
on quality grounds. For example, a disallowed charge state or modification. If a ratio is negative, which indicates
some instrument or peak detection problem, (or an inappropriate correction),
this is reported. However, a negative ratio is discarded when calculating the protein ratio.
The screen shot illustrates a Select Summary report, where detailed information is only displayed for the strongest
match to each unique peptide sequence. The quantitation information also pertains to just the strongest match. It is
not an average of all the matches in the row.
There are a number of switches to control the presentation of quantitation results. The default values are set in the quantitation method
and can be changed in an
individual report using the format controls,
or by appending the relevant switch to the report URL. Switches take the form label=value and
the delimiter between switches is an ampersand (&). For example, if the report URL was:
http://local-server/mascot/cgi/master_results.pl?file=../data/20040121/F001847.dat
The protein ratio type could be changed by appending "_quant_protein_ratio_type=average":
http://local-server/mascot/cgi/master_results.pl?file=../data/20040121/F001847.dat&_quant_protein_ratio_type=average
Labels and values are not case sensitive. Note that many labels begin with an underscore character. Values that are not
literal strings are shown in italics.
| URL |
Value |
Description |
| _quant_protein_ratio_type |
average |
The protein ratio is the geometric mean of the peptide ratios |
| median |
The median peptide ratio is selected to represent the protein ratio |
| weighted |
For each component, the intensity values of the assigned peptides are summed and the
protein ratio(s) calculated from the summed values. |
| _quant_norm_method |
none |
No normalisation is performed |
| average |
For each peptide ratio, a correction factor is applied such that the geometric mean of the
ratios for all peptide matches that pass the quality tests is unity |
| median |
For each peptide ratio, a correction factor is applied such that the median of the ratios
for all peptide matches that pass the quality tests is unity |
| sum |
A correction factor is applied such that the sum of the intensities for a reporter ion peak
over all peptide matches that pass the quality tests is the same for all the reporter ions |
| _quant_report_detail |
1 |
Set to 1 to display ratios for individual peptides |
| _quant_outliers_method |
none |
No outlier removal |
| auto |
Use Dixon's method if the number of values is between 4 and 25, Rosners if 26 or more |
| dixons |
Outlier removal using Dixons method |
| grubbs |
Outlier removal using Grubbs method |
| rosners |
Outlier removal using Rosners method |
| _quant_min_num_peptides |
N |
The minimum number of valid peptide match ratios that are required in order to report a protein hit ratio |
| _min_precursor_charge |
N |
A peptide ratio will not be reported if the precursor charge is less than this value |
| _quant_pep_threshold_type |
minimum score |
Threshold for calculating a peptide ratio is a minimum score (value specified in _quant_pep_threshold_value) |
| maximum expect |
Threshold for calculating a peptide ratio is a maximum expect (value specified in _quant_pep_threshold_value) |
| at least identity |
Threshold for calculating a peptide ratio is at least identity |
| at least homology |
Threshold for calculating a peptide ratio is at least homology |
| _quant_pep_threshold_value |
N |
Value for threshold type specified in _quant_pep_threshold_type |
| _quant_unique_pepseq |
1 |
If set to 1, ratios are only reported for peptide sequences that are unique to one protein hit, |
|