Sequence Database Setup: NCBI EST
| These are Predefined Database Definitions |
| The information on this page is maintained as a service to users of Mascot 2.3 and earlier.
In Mascot 2.4, the NCBI EST divisions are predefined databases, meaning up-to-date configuration information can be
downloaded automatically by Mascot Database Manager.
|
|
| EST_others |
| As of April 2012, the compressed EST_others file from NCBI was 11 GB
and the unpacked Fasta was 40 GB. Address space constraints
mean that it is no longer possible to memory map this file or to build a taxonomy index using 32-bit
executables. Even so, given sufficient disk space,
you should still be able to search EST_others on a 32-bit system, provided you configure without
memory mapping or taxonomy.
NCBI have no plans to split EST_others, so a more practical alternative is the set of 10
EMBL EST files,
which are more evenly divided. As of April 2012, the largest (plants) was 4.3 GB compressed and 17 GB unpacked.
If you only require sequences for a particular organism, another possibility is to download an
organism specific EST database.
|
|
Overview
Three EST databases are compiled by the
NCBI (National Center for Biotechnology Information). They
contain "single-pass" cDNA sequences, or Expressed Sequence Tags, from the EST divisions of
GenBank.
There are currently three EST databases: human, mouse, and others. This document uses the "others" database
as an example. To work with the human or mouse databases, simply substitute the word "human" or
"mouse" for "others". For example, the human
compressed Fasta file is est_human.gz, the db_update.pl keyword is EST_human_from_NCBI, the recommended
Mascot name is EST_human, etc.
Download
ftp://ftp.ncbi.nih.gov/blast/db/FASTA/est_others.gz
for the latest release.
Taxonomy
Taxonomy for NCBI EST databases is predefined in mascot.dat. For EST_others, choose
"dbEST FASTA using GI2TAXID". (There is no value in building taxonomy indexes
for human or mouse because these are single organism
databases.) The following taxonomy files are required:
ftp://ftp.ncbi.nih.gov/pub/taxonomy/gi_taxid_nucl.dmp.gz
ftp://ftp.ncbi.nih.gov/pub/taxonomy/taxdump.tar.gz
Note that the taxonomy files go into the taxonomy directory, not into the sequence database
directory. Also, some files need to be unpacked (using tar) as well as uncompressed.
Unigene
The NCBI UniGene
indexes are created by automatically partitioning GenBank sequences into non-redundant sets of
gene-oriented clusters. If UniGene indexes are available locally, results from Mascot searches of
EST databases can be grouped and reported by gene family, rather than by raw EST accession numbers.
In Mascot 2.4, the following UniGene indexes are included in the predefined database definitions.
To enable UniGene indexes in earlier versions, refer to your local copy of this help page
- EST_human
- EST_mouse
- EST_others
- Anopheles_gambiae
- Arabidopsis_thaliana
- Bos_taurus
- Caenorhabditis_elegans
- Chlamydomonas_reinhardtii
- Danio_rerio
- Dictyostelium_discoideum
- Drosophila_melanogaster
- Hordeum_vulgare
- Oryza_sativa
- Rattus_norvegicus
- Takifugu_rubripes
- Triticum_aestivum
- Xenopus_laevis
- Zea_mays
Parse Rules
A typical Fasta title line is:
>gi|16764|emb|Z17609.1|Z17609
ATTS0183 Gif-SeedA+B Arabidopsis thaliana cDNA clone YAP043T 3'
The gi number is the most reliable identifier. Suitable parse rules are:
Accession from Fasta title: ">\(gi|[0-9]*\)"
Description from Fasta title: ">[^ ]* \(.*\)"
If an entry in EST_others represents multiple source database entries, the Fasta title lines are concatenated
together with CTRL+A as the delimiter.
Configuration (Mascot 2.3 and earlier)
For this example, est_others.gz was downloaded to a folder named
C:\Inetpub\MASCOT\sequence\EST_others\current.
The file was decompressed using gzip,
and renamed to EST_others_20110601.fasta.
Taxonomy and memory mapping have been enabled, which is only possible for EST_others when using 64-bit Mascot.
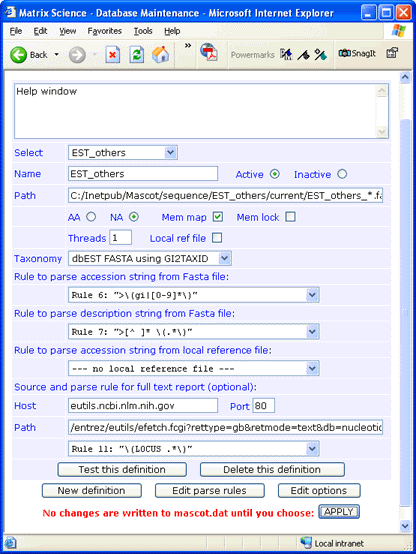
There is no downloadable full text file for EST_others, but full text for individual entries can be retrieved across the web
from the NCBI Entrez server. The syntax
for the Path field is:
/entrez/eutils/efetch.fcgi?rettype=gb&retmode=text&db=nucleotide&tool=mascot&id=#ACCESSION#
If you don't require full text in a Mascot Protein View report, simply leave the Host, Port, and Path fields blank
and choose
--- no full text report ---
in the drop down list.
Always test a new definition before applying the changes to mascot.dat.
|