Sequence Database Setup: Generic Database
Overview
A very simple configuration is sufficient for the common case of a sequence database where all the entries have the same taxonomy and
there is no full text reference file. To add such a database to Mascot, the requirements are:
- A local copy of the database in Fasta format
- Each sequence must have a unique identifier (accession string)
Download a Fasta File
Mascot can search both protein and nucleic acid sequences. For a PMF search,
the database should be either protein sequences or nucleic acid sequences
equivalent to proteins, such as mRNA sequences. For a search of MS/MS data, the sequences can also be EST or genomic DNA data.
The relative merits of searching protein, EST and DNA sequences are discussed in
TRENDS in Biotechnology 19(10)
S17-S22(2001).
In all cases, a local copy of the database in Fasta format is required. A suitable file can usually be
downloaded from the NCBI web site, and a good place to start is
the NCBI taxonomy browser.
Protein Database Example
See also UniProt Proteomes.
We'd like a protein database for Chinese hamster (Cricetulus griseus), because this
important animal is not yet well represented in SwissProt.
In the NCBI taxonomy browser,
search for Chinese hamster as a complete name.
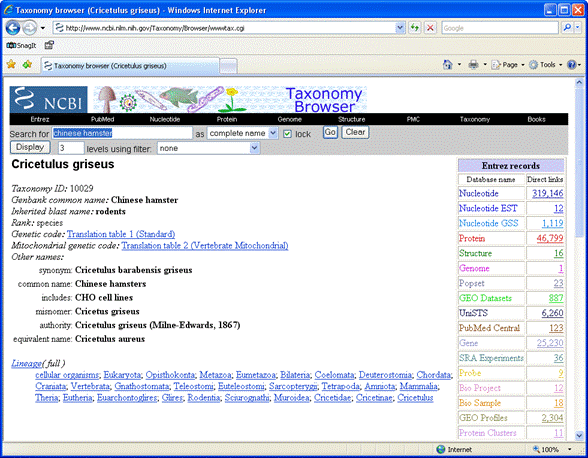
The table of Entrez records shows that there are 46,799 protein entries. Follow the link
to the first page of a listing of these entries. From the Send to drop-down list,
choose File and Fasta format
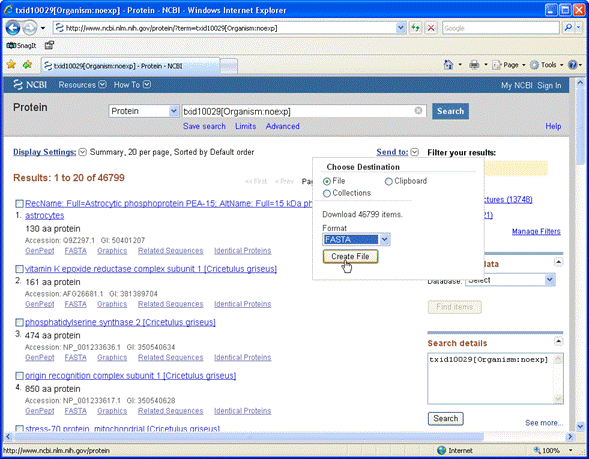
This is a fasta file suitable for searching with Mascot. For a protein Fasta file downloaded
from NCBI, create a new custom definition using NCBI_AA_template as the template.
In Mascot 2.3 and earlier, configure as shown below.
EST Database Example
We're working on a disease that affects oranges, but this fruit is very
poorly represented in the protein databases.
In the NCBI taxonomy browser,
search for citrus as a complete name. There are several
oranges in the list, but individual strains, such as Seville orange, have very few sequences
of any type.
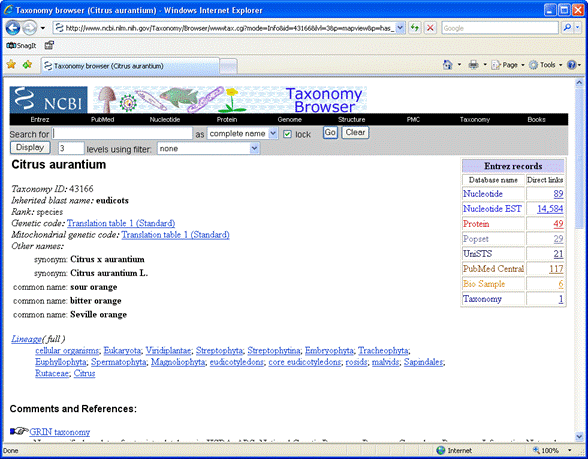
Going up one level, to citrus, is more fruitful, showing more than
half a million EST sequences. Follow the Nucleotide EST link
to the first page of a listing of these entries. From the Send to drop-down list,
choose File and Fasta format

This is a fasta file suitable for searching with Mascot. For a nucleic acid Fasta file
downloaded from NCBI, create a new custom definition using NCBI_NA_template as the template.
In Mascot 2.3 and earlier, configure as shown below.
Genome Database Example
Our organism of interest
is Helicobacter pylori and we want to search the genome of a particular strain, HPAG1.
In the NCBI taxonomy browser,
search for Helicobacter pylori as a complete name.
Follow the link for Helicobacter pylori HPAG1.
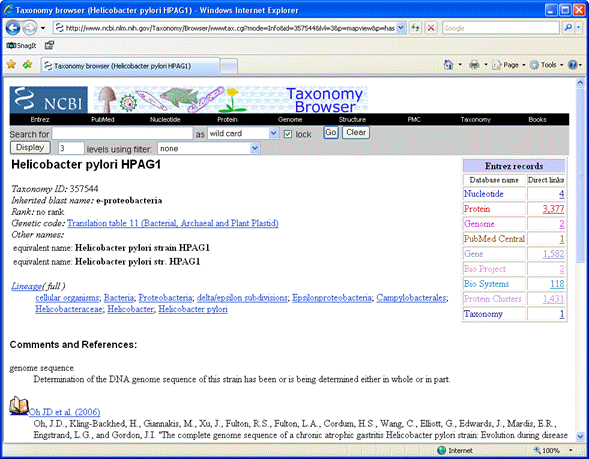
There are many routes to get to the genome sequence data and there is a choice of assemblies. In this particular
case, following the Nucleotide link in the table of Entrez records is the most direct route. This lists
four sequences, two for the plasmid and two for the chromosome. We might decide to select the NCBI Reference Sequences
for the chromosome and plasmid.
From the Send to drop-down list, choose File and Fasta format
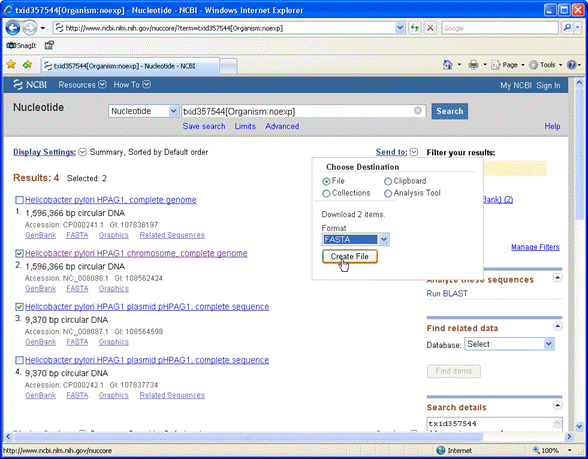
The assembled
chromosome is a single sequence of length 1,596,366 bases. This is not ideal for a Mascot search, because it would make the reports
very unwieldy. For efficient searching, genomic DNA needs to be split into shorter segments, with small overlaps
to ensure no peptides are lost because they span a break. Ideally,
you also want to maintain the original forward and reverse frame numbering from segment to segment. A simple Perl script to split
a long sequence can be downloaded here. Usage information can be obtained by
executing it with no arguments.
Note: We have had several reports that this file is unpacked automatically
when downloaded using Microsoft Internet Explorer on a Windows PC. If you cannot
open the file in Winzip, try to open it in a text editor like WordPad. If it looks
like text, then it has been unpacked, and you only need to rename the file to splitter.pl.
The output of the splitter is a fasta file suitable for searching with Mascot. For
genome sequences that have been split
into segments, create a new custom definition using simple_NA_template as the template.
In Mascot 2.3 and earlier, configure as shown below.
Parse Rules
Every entry in the database must have a unique accession string.
Mascot Database Manager
makes it easy to select suitable parse rules by displaying examples of what would be matched
by the available rules.
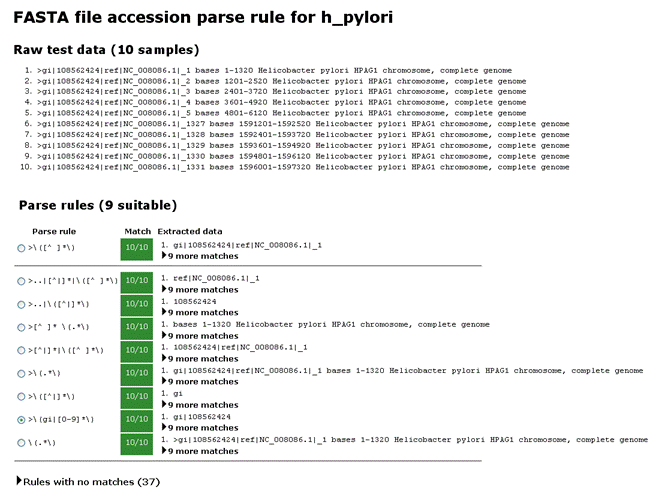
Uniprot (SwissProt and Trembl) and NCBI (Genbank) have relatively complicated title lines, but parse rules for these are pre-defined
in Mascot. In most other cases, a simple rule that takes everything between the ">" symbol
and the first space as the accession will work.
Everything after the first space can be treated as the description. These are the rules used in
Database Manager predefined definitions simple_AA_template and simple_NA_template.
">\([^ ]*\)"
">[^ ]* \(.*\)"
In the unlikely event that you need to create a new parse rule:
- If a rule looks like it should work, and doesn't, it may be because the space is actually a tab. If
this is the case, then you can use a character class that includes or excludes all the printing characters
">\([!-~]*\)"
">[!-~]*[^!-~]\(.*\)"
- Don't make a parse rule more precise than it needs to be. It is more likely to go wrong if there is
some change to the Fasta title syntax.
- Mascot parse rules are Basic Regular Expressions, as used in grep, not Extended Regular Expressions,
as used in Perl.
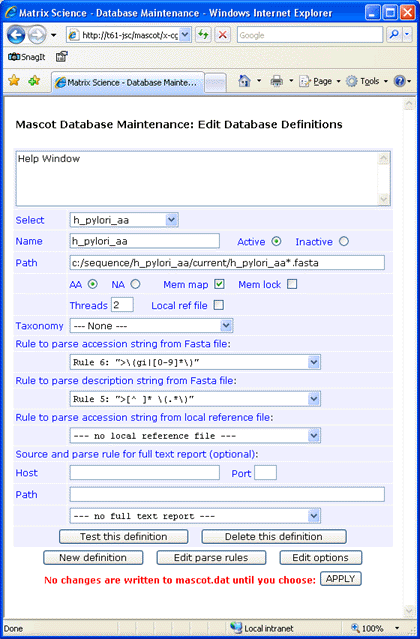
As a first example, protein sequences for Helicobacter pylori were downloaded from NCBI to a folder named
C:\sequence\h_pylori_aa\current and the Fasta file renamed to h_pylori_aa.fasta.
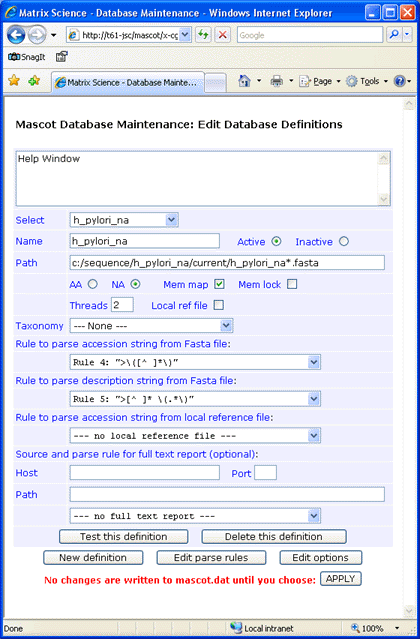
As a second example, the plasmid and chromosome sequences for Helicobacter pylori were downloaded from NCBI to a temporary file.
The splitter utility was used to divide the sequences into chunks of 12000 bases. The new file was renamed
h_pylori_na.fasta and moved to C:\sequence\h_pylori_na\current.
Tips:
- Make sure you set the AA / NA radio button correctly first time
- Databases should be memory mapped but not memory locked
- The wild card in the database path is required
- The value of threads should be set to the number of processor
cores up to a maximum of 4 times the number of processors licensed for Mascot.
- Always test a new definition before applying the changes to mascot.dat
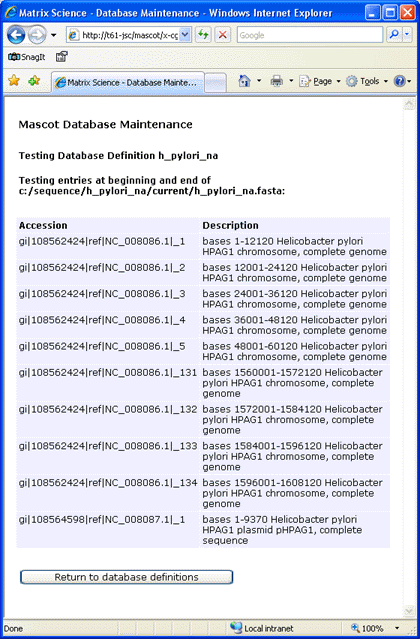
Assuming that there are no error messages from testing the database, choose Apply to save the new
configuration in mascot.dat. Then, follow the link to Database Status and verify that there are no
errors when the new database is compressed and tested. Once the database status reads "In Use",
the database is available for searching.
|