Sequence Database Setup: nr
| This is a Predefined Database Definition |
| The information on this page is maintained as a service to users of Mascot 2.3 and earlier.
In Mascot 2.4, NCBInr is a predefined database, meaning up-to-date configuration information can be
downloaded automatically by Mascot Database Manager.
|
|
Overview
The nr database is compiled by the
NCBI (National Center for Biotechnology Information) as a
protein database for Blast searches. It
contains non-identical sequences from
GenBank CDS translations,
PDB,
Swiss-Prot,
PIR, and
PRF.
The strengths of nr are that it is comprehensive and updated very frequently.
Download
ftp://ftp.ncbi.nih.gov/blast/db/FASTA/nr.gz
for the current release.
To download NCBInr updates automatically in Mascot 2.3 and earlier,
the relevant definition block in db_update.pl is NCBInr_from_NCBI.
Taxonomy
Taxonomy for nr is predefined in mascot.dat, choose "NCBI nr FASTA using GI2TAXID".
The following taxonomy files are required:
ftp://ftp.ncbi.nih.gov/pub/taxonomy/gi_taxid_prot.dmp.gz
ftp://ftp.ncbi.nih.gov/pub/taxonomy/taxdump.tar.gz
Note that the taxonomy files go into the taxonomy directory, not into the sequence database
directory. Also, some files need to be unpacked (using tar) as well as uncompressed.
Parse Rules
A typical Fasta title line is:
>gi|21305377|gb|AAM45611.1|AF384285_1
(AF384285) envelope protein [Human immunodeficiency virus type 1]
The gi number is the most reliable identifier. Suitable parse rules are:
Accession from Fasta title: ">\(gi|[0-9]*\)"
Description from Fasta title: ">[^ ]* \(.*\)"
If an entry in nr represents multiple source database entries, the Fasta title lines are concatenated
together with CTRL+A as the delimiter.
Configuration (Mascot 2.3 and earlier)
| NCBInr failing to come on-line after an update |
| NCBInr has grown so large that Mascot running on a 32-bit operating system
encounters memory issues when processing it and crashes. The fix is to edit mascot.dat
change
IgnoreDupeAccessions EST_others
to
IgnoreDupeAccessions EST_others NCBInr
This means that, if there are duplicate accessions in the
database, matches will be misreported. NCBInr has been
very reliable, and we are not aware of this ever happening,
but we do not recommend adding other very large databases to the
IgnoreDupeAccessions directive unless you can verify by other
means that there are no duplicate accessions.
|
|
For this example, nr.gz was downloaded to a folder named
C:\Inetpub\MASCOT\sequence\NCBInr\current.
The file was decompressed using gzip,
and renamed to NCBInr_20020601.fasta.
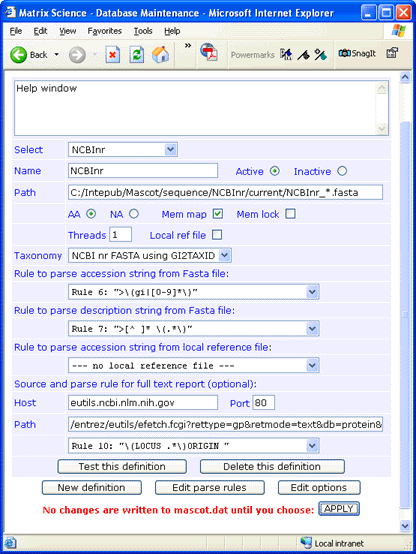
There is no downloadable full text file for nr, but full text for individual entries can be retrieved across the web
from the NCBI Entrez server. The syntax
for the Path field is:
/entrez/eutils/efetch.fcgi?rettype=gp&retmode=text&db=protein&tool=mascot&id=#ACCESSION#
If you don't require full text in a Mascot Protein View report, simply leave the Host, Port, and Path fields blank
and choose
--- no full text report ---
in the drop down list.
Always test a new definition before applying the changes to mascot.dat.
|