Sequence Database Setup: SwissProt
| This is a Predefined Database Definition |
| The information on this page is maintained as a service to users of Mascot 2.3 and earlier.
In Mascot 2.4, SwissProt is a predefined database, meaning up-to-date configuration information can be
downloaded automatically by Mascot Database Manager.
Choose SwissProt_ID to use the ID as the unique
identifier or choose SwissProt_AC to use the AC.
|
|
Overview
UniProtKB/Swiss-Prot
(reviewed) is a high quality manually annotated and non-redundant protein sequence database,
which brings together experimental results, computed features and scientific conclusions.
About 85 % of the protein sequences in UniProtKB are derived from the translation
of coding sequences (CDS) from the EMBL-Bank/GenBank/DDBJ public nucleic acid
databases.
UniProtKB is a collaboration between the
European Bioinformatics Institute, the
Swiss Institute of Bioinformatics and the
Protein Information Resource.
Download
Expasy:
ftp://ftp.uniprot.org/pub/databases/uniprot/current_release/knowledgebase/complete/
EBI:
ftp://ftp.ebi.ac.uk/pub/databases/uniprot/knowledgebase
The EBI site mirrors the Expasy site. The relevant files are:
- Version info: reldate.txt
- SwissProt Fasta file: uniprot_sprot.fasta.gz
- SwissProt Dat file: uniprot_sprot.dat.gz
To download SwissProt updates automatically in Mascot 2.3 and earlier,
the relevant definition block in db_update.pl is SwissProt_complete_from_EBI.
There is also a definition for downloading just the SwissProt
Fasta file: SwissProt_fasta_only_from_EBI.
Taxonomy
Taxonomy is predefined in mascot.dat. Even if you have the SwissProt Dat file,
choose "SwissProt FASTA".
In Mascot 2.3 and earlier, verify that the taxonomy definition in mascot.dat is up to date:
# TAXONOMY FOR SwissProt or Trembl from the fasta file
Taxonomy_3
Identifier SwissProt FASTA
Enabled 1 # 0 to disable it
FromRefFile 0
DescriptionLineSep 0 # ctrl a - hex code '1'. For multiple descriptions per entry
SpeciesFiles NCBI:names.dmp, SWISSPROT:speclist.txt
NodesFiles NCBI:nodes.dmp, NCBI:merged.dmp
DefaultRule SWISSPROT, CHOP: ">[^_]*_\([^ ]*\) " # Anything after _ before space
end
#
Note that mascot.dat must be saved as plain text, so be careful if using a word processor,
and ensure the filename is not changed to mascot.dat.txt or something.
The following taxonomy files are required:
ftp://ftp.ncbi.nih.gov/pub/taxonomy/taxdump.tar.gz
ftp://ftp.ebi.ac.uk/pub/databases/uniprot/knowledgebase/docs/speclist.txt
Taxonomy files go into the taxonomy directory, not into the sequence database
directory. Also, some files need to be unpacked (using tar) as well as uncompressed.
| SwissProt Release 2011_06 in Mascot 2.3 and earlier |
| A change in the format of speclist.txt
broke taxonomy assignment in Mascot 2.3 and earlier for SwissProt Release 2011_06 onwards.
The symptom is that searches using a taxonomy filter return no matches.
A modified file that fixes the problem can be downloaded from this URL:
speclist.txt.
If you use the database update script (db_update.pl) to perform automatic updates
of SwissProt, change the URL for downloading speclist.txt in the relevant definition
block to http://www.matrixscience.com/downloads/speclist.txt
If you have discovered this problem after updating Trembl, the procedure
to correct it is as follows:
- Windows: stop the Mascot service, Unix: kill ms-monitor.exe
- Delete the *.stats file in the database current directory
- Download the modified speclist.txt to the taxonomy directory
- Windows: start the Mascot service, Unix: execute ms-monitor.exe
|
|
Parse Rules
A typical SwissProt Fasta title line is:
>sp|Q4U9M9|104K_THEAN 104 kDa microneme/rhoptry antigen OS=Theileria annulata GN=TA08425 PE=3 SV=1
You can use either the ID (104K_THEAN) or
the AC (Q4U9M9) as the identifier.
Many people prefer the ID because it is semi-descriptive.
ID from Fasta title: ">..|[^|]*|\([^ ]*\)"
AC from Fasta title: ">..|\([^|]*\)"
Description from Fasta title: ">[^ ]* \(.*\)"
The corresponding lines in the Dat file are:
ID 104K_THEAN Reviewed; 893 AA.
AC Q4U9M9;
ID from Ref file: "^ID \([^ ]*\)"
AC from Ref file: "^AC \([-A-Z0-9_]*\)"
Configuration (Mascot 2.3 and earlier)
For this first example, the database files were downloaded to
C:\Inetpub\MASCOT\sequence\SwissProt\current,
decompressed using gzip,
and renamed to SwissProt_56.0.dat and SwissProt_56.0.fasta.
When updating an active database, it is important to rename the Fasta file last, because Mascot
will begin database exchange as soon as it sees a new Fasta file that matches the wildcard path for
the database.
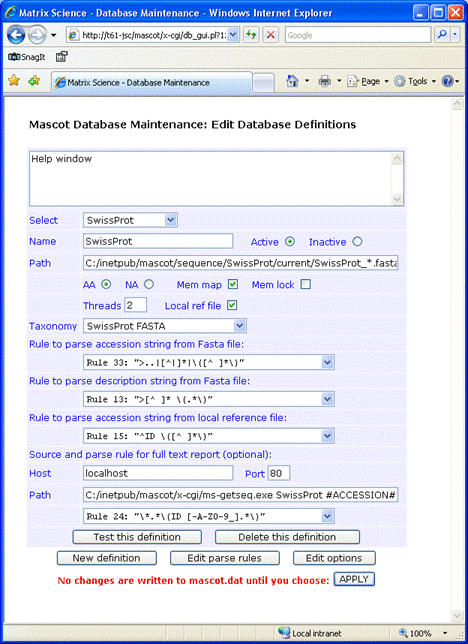
If you decide not to have the reference file locally, full text for individual entries can be retrieved across the web
from Uniprot or an SRS server.
For Uniprot, the required entries are:
Host: www.uniprot.org
Port: 80
Path: /uniprot/#ACCESSION#.txt
Parse rule: RULE_23 "\(.*\)"
Where #ACCESSION# represents either the AC or ID. For an SRS
server, the syntax for the Path field is:
Retrieve by ID: /srsbin/cgi-bin/wgetz?-e+[SWISSPROT-id:#ACCESSION#]+-vn+2
Retrieve by AC: /srsbin/cgi-bin/wgetz?-e+[SWISSPROT-acc:#ACCESSION#]+-vn+2
This screen shot illustrates a configuration in which the identifier is AC, there is no local Dat file,
and full text is retrieved from an SRS server:
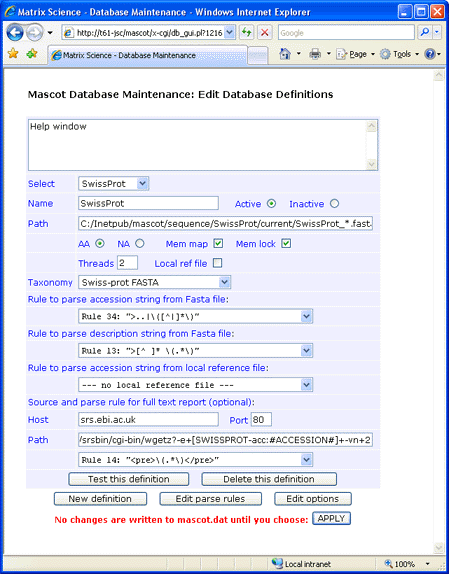
Make sure that the final parse rule has the correct case. Early versions of wgetz return HTML pages tagged
with <PRE>, while later versions use <pre>. Parse rules are always case sensitive.
If you don't require full text in a Mascot Protein View report, simply leave the Host, Port,
and Path fields blank and choose
--- no full text report ---
in the drop down list.
Always test a new definition before applying the changes to mascot.dat.
|