Thermo Xcalibur
There is a bewildering choice of software to convert Xcalibur data into peak lists and
submit these to a Mascot Server for searching. This page lists some of the more widely used options.
Remember that the spectra in Xcalibur raw files may
contain centroid data, rather than profile data. With the latest hybrid instruments, it has
become common practice to save high resolution survey scans as profile and low resolution MS/MS scans as centroid.
Mascot Distiller can be used to browse
Xcalibur raw files, and process them into high quality peak
lists that can be saved or submitted direct to a Mascot Server for searching.
With the appropriate Distiller Toolboxes, the search results can be imported back into Distiller for
further examination or used as the basis for quantitation. If the optional Mascot Daemon Toolbox is
installed, these processes can be automated using Mascot Daemon.
If your MS/MS data is centroided, you can choose to create a peak list direct from the
centroid values already present in the raw file.
This is extremely fast and the peak list is fine for most purposes. Choose extract_msn.opt as the processing
options when opening the raw file as a new project.
With high resolution data from an FT or Orbitrap, you may wish to take a little longer, and peak pick
the survey scans, so as to obtain more reliable detection of the 12C peaks. For high charge state
data, you may wish to peak pick the MS/MS scans so that the peaks can be de-isotoped and de-charged. Mascot only tries
to match 1+ and 2+ fragments, so de-charging to 1+ becomes important when the precursor is 4+ or higher. Full details of how to select
and modify the processing options can be found in the Distiller help file, (see especially the 'More about peak picking' topic in
the Reference chapter).
Besides the quality of the peak lists, the other advantages of using Mascot Distiller are that it provides a
universal interface to other raw file formats, and it is fully integrated with Mascot Server and Mascot Daemon.
You can, for example, use Mascot Daemon to process batches of files automatically, saving Distiller project files that
contain both the peak lists and the search results.
Note: To open Xcalibur 2.1 files in Distiller 2.3, install MSFileReader after installing Distiller.
Mascot Daemon can be used to process batches
of RAW files by choosing either Mascot Distiller or ThermoFinnigan LCQ / DECA RAW file
as the data import filter.
Mascot Distiller is the more powerful option, and this is the required route if you intend to use
Distiller for quantitation. Distiller requires the optional Mascot Daemon Toolbox to allow the Distiller libraries
to be called from Mascot Daemon. When this toolbox is active, Mascot Distiller will appear
automatically on the list of data import filters in Daemon.
If you choose ThermoFinnigan LCQ / DECA RAW file, Daemon executes
extract_msn to
convert each raw file into a set of DTA files, then merges these into an MGF file. Unlike the
lcq_dta web browser
form, Daemon executes extract_msn on the Daemon PC, so this option is
available even if your Mascot server is on a Unix platform.
If you have a set of DTA format peak lists, but no raw file, you can also use Daemon to merge the DTAs
into an MGF file for searching. Select the DTA files in Windows Explorer and drag and drop them into the Daemon
data files list box on the Task Editor tab. Then, check the box for Merge MS/MS files into a single search.
Real-time monitor
By running Mascot Daemon in real-time monitor mode, each RAW file can be searched automatically,
as soon as acquisition is complete. First, create a suitable parameter set for the task:
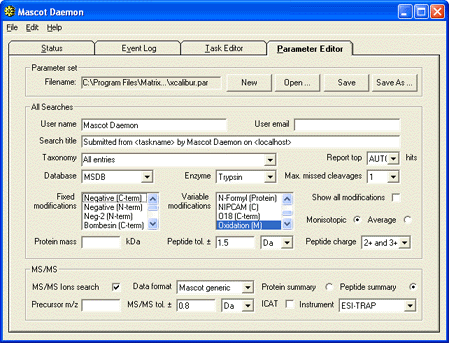
(Note that the file format is Mascot Generic, not DTA, because Daemon data import filters
always create MGF files.) Second, create a real-time monitor task to monitor the directory where the RAW files
are being created. Remember to select the correct parameter file, and choose either Mascot Distiller or
ThermoFinnigan LCQ / DECA RAW file (to use extract_msn) as the data import filter.

The data import filter processing options are specified by choosing the Options button next to the
data import filter list box. For Distiller, you may have something like this
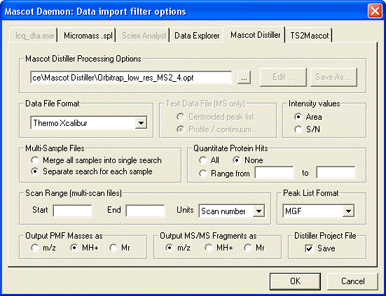
For extract_msn, these would be typical settings:

Troubleshooting
- The most recent version of extract_msn changes the name of the executable to ExtractMSn.exe.
Daemon 2.3 and earlier are not aware of the new executable name, and will not accept it.
Make a copy of the executable and rename it to extract_msn.exe, then browse to this file to select
it in the Daemon preferences dialog
-
In real-time monitor mode, it is important that Mascot Daemon waits until acquisition is complete before
processing the RAW file into peak lists. To avoid taking a file while it is still being written, Daemon checks the file size
at intervals, and waits until it has stopped increasing. The default interval is 60 seconds, which
may not be long enough when the file size grows only slowly. If Daemon tries to process a RAW file
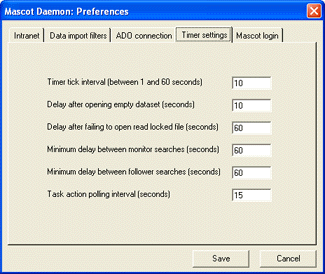
before acquisition is complete, increase this interval by going to the
Timer Settings tab of the Preferences dialog. Increase the value of 'Delay after failing to open
read-locked file' until the problem disappears.
- If there are problems processing very large RAW files, check that you have adequate disk space. When Daemon
processes a RAW file using extract_msn, the workspace is in the local user's temp
directory, the location of which is system dependent. Under Windows 2000 and later, the path is
C:\Documents and Settings\<Windows User Name>\Local Settings\Temp. You'll know when you've found the right
location because it will contain a sub-directory called Mascot_Daemon_workspace.
- If Mascot Daemon reports "No output from lcq_dta.exe (check parameters)"
or the lcq_dta shell form returns "Must choose at least one query for repeat search"
this means that no DTA files were produced. The most common causes are (i) the extract_msn parameters
are too restrictive, (ii) the data file does not contain MS/MS scans, (iii) the version of extract_msn
is older than the version of Xcalibur used to create the data file. The easiest way to investigate
and debug this problem is to execute extract_msn at a command prompt, using identical processing
parameters.
- If your Mascot server runs under Windows XP, and you get the message "cannot create
temporary directory" when you try to use the lcq_dta shell form, this may be because
the security settings do not allow CGI programs to execute the command processor. A fix
is described on the Support page,
in the Windows XP section.
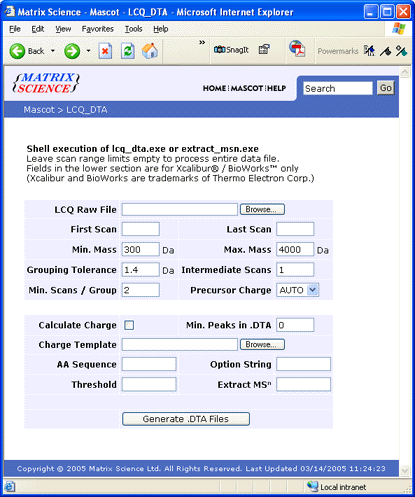
If you have a Windows-based Mascot server in-house, you can use the lcq_dta shell search form
to upload and process the RAW file. When this form is submitted, the processing options are passed
to extract_msn running on the server. The RAW file is processed into DTA files which are automatically
merged into a single file, pre-loaded into a Mascot search form.
When Mascot is first installed, you need to edit the underlying Perl script (lcq_dta_shell.pl) to
specify the locations of a workspace directory and the
directory containing the extract_msn executable. These are
defined by two variables near the top of the script:
# local name of temp directory on Mascot server (no trailing slash)
my $tempDir = "c:\\temp";
# local path to lcq_dta.exe or extract_msn.exe on Mascot server
my $lcqExe = "c:\\Xcalibur\\System\\Programs\\extract_msn.exe";
Note the use of double backslashes in the path names.
Note: If you are submitting searches to the public web site, remember
that the size of the upload file is limited to 1200 spectra. To avoid these limits, license Mascot
to run on your in-house server.
Support for submitting searches direct to a Mascot Server was added to Thermo's
Bioworks in version 3.2, but we advise using
Bioworks 3.3 SP1 to avoid some known issues with the first release.
Mascot Server must be version 2.1 or later. In Bioworks browser, choose Configuration off the Options menu.
In the dialog, select Mascot Search and enter the Mascot Server URL in the form http://ec-vm2/mascot/cgi
where ec-vm2 is replaced by the hostname of your local server.
When a data file is loaded, you can choose Mascot off the Actions menu to submit a search. Bioworks
creates and saves an mzData format peak list for submission to Mascot.
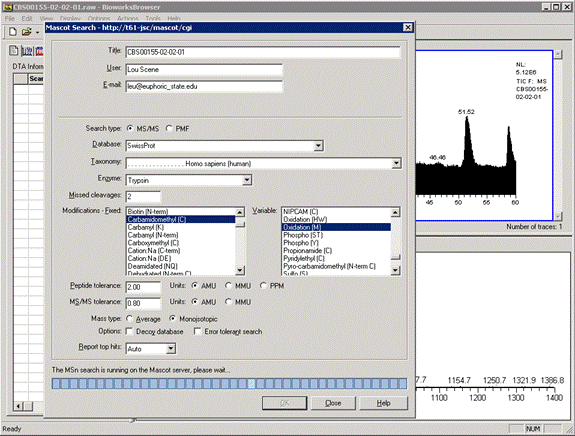
When the search is complete, you can load the Mascot results report in a web browser or download the results
file to the Bioworks PC. Note that Bioworks has been superceded by Proteome Discoverer, and is no longer available.
Thermo's Proteome
Discoverer provides fully automated raw file processing and search submission. Peak picking and
search parameters are selected in a workflow wizard. When the search is complete, the results are imported into
Proteome Discoverer, where they can be filtered and inspected.
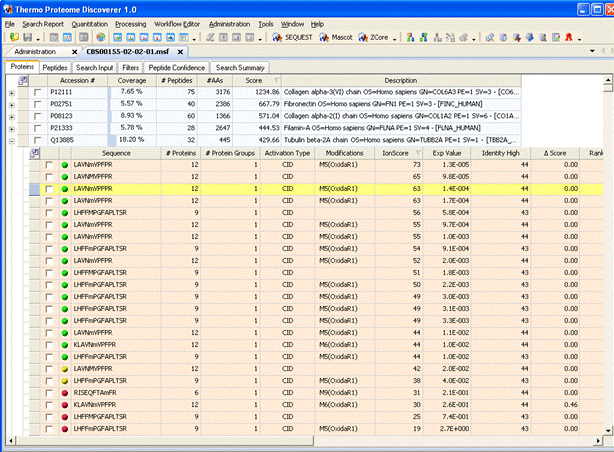
(Note that the local Mascot Server URL must be entered in the form http://ec-vm2/mascot/
where ec-vm2 is replaced by the hostname of your local server.)
Utilities
Mascot supports the Sequest DTA peak list format. However, if the data are from an LC-MS/MS experiment,
searching individual DTA files is inefficient, and doesn't allow Mascot to generate a proper results summary.
You can concatenate a set of DTA files into an MGF peak list using one of these utilities:
- merge.pl, a Perl script (any platform)
- merge.bat, a DOS batch file (Windows)
- merge.sh, a shell script (Unix)
Download all three utilities for Windows
or Unix
If possible, you should choose the Perl script, because this creates a Mascot Generic Format (MGF)
file in which each DTA file name is preserved as a spectrum title. This makes it easier to compare the Mascot
search results with the original data, because you can identify the scan range represented by each
spectrum. It also enables the origin of each DTA file to be tracked when data from
multiple RAW files from a MudPIT experiment are merged together.
Most Unix systems will already have Perl installed. If your Windows system doesn't have Perl, it
can be downloaded free from ActiveState.
(Quote from Bugzilla:
"Any machine that doesn't have Perl on it is a sad machine indeed.")
The original Windows console (DOS) utility for converting a raw file into a set of DTA format peak lists was
developed by John Yates' group at U. Washington and called extractms. When first included with Xcalibur, it was called
lcq_dta.exe. Over the years, the name changed to extract_msn.exe and it
became a component of Thermo's Bioworks application package. With version 5, the executable became extract_msn_com.exe.
In 2011, it was renamed to ExtractMSn.exe and gained an optional GUI.
To avoid repetition, we will refer to all versions of this utility as extract_msn.
In general, you cannot process raw files
from one release of Xcalibur using extract_msn from an earlier release. Unfortunately, it isn't always
easy to figure out which version you have, and all versions depend on a changing population of dynamic link libraries (DLLs).
Usage information can be displayed by executing extract_msn without
any arguments. This is also a quick way to tell whether the required DLLs are
present and correct. Additional information can be found in your Xcalibur or Bioworks documentation.
The following are worth noting:
- Intermediate scans (-S): Although it looks like it should be OK to set S to zero, this
can sometimes result in no output
- Min. Peaks in DTA (-I): The default is 0, but this should always be set to a sensible number,
say 10, to remove empty or near empty scans, since these can never give significant matches
in Mascot.
- Precursor Charge (-C): With triple-play data, precursor charge state determination is
fairly sophisticated, and the default settings should not be changed. If your data don't include zoom scans, the code
attempts to recognise singly charged precursors, while precursors with higher charge states are
output twice, with 2+ and 3+ charge states.
- TIC Threshold (-E): Not described in the Usage information
- Extract MSn (-P): Not described in the Usage information
Mascot supports the DTA format. However, if the data are from an LC-MS/MS experiment,
searching individual DTA files is inefficient, and doesn't allow Mascot to generate a proper results summary.
If you have a set of DTA files, it will usually be best to merge them into a single file.
If you have Mascot in-house, you can have Mascot Daemon take care of this, automatically.
If you want to use extract_msn on a different PC from the one where Xcalibur and Bioworks are installed,
extract_msn ver. 5.0
can be downloaded from Thermo's
customer download area.
You will also need to install MSFileReader to provide the supporting libraries.
Note: extract_msn does not perform centroiding of profile data. If you generate DTA files from a
RAW file containing profile data, the DTA files are themselves profile data. Zero intensity values are dropped,
and non-zero intensities are output at 0.1 Da intervals. Mascot deals with this as best it can by performing simple
peak detection, but this is less than ideal. The other problem of working with profile data is
that the DTA files will be very large, and you may
occasionally get a Mascot error message that there are more than 10,000 data points in a single spectrum.
To open Xcalibur 2.1 files in Mascot Distiller 2.3, you must also install Thermo's
MSFileReader utility.
This is a standalone installation of XRawfile2.dll, which permits programmatic access of Thermo data files via a COM interface.
Note: MSFileReader must be installed after Mascot Distiller 2.3. If you
subsequently reinstall Distiller, you must then repair or reinstall MSFileReader.
DeconMSn has been developed at
Pacific Northwest National Laboratory. It requires Xcalibur and Microsoft .NET 1.1 or later to be installed. It is
not clear whether it can be made to run stand-alone, on a system without a full installation of Xcalibur.
DeconMSn can output either DTA or MGF peak lists.
With high resolution data, parent monoisotopic mass is calculated
using a modified THRASH approach. For low-resolution data, DeconMSn uses a support-vector machine based
charge-detection algorithm to determine parent mass.
DTASuperCharge
is a component of MSQuant. It creates MGF
peak lists from raw files, retaining the retention time and scan number information required by MSQuant.
It requires Xcalibur (including the XDK) and Microsoft .NET 2.0 or later to be installed.
It is not clear whether it can be made to run stand-alone, on a system without a full installation of Xcalibur.
Raw2MSM
creates MGF peak list files from Xcalibur raw files, and works best with high accuracy LC-MS/MS data, from an Orbitrap
or FT instrument. For some mysterious reason, the MGF files are given the extension MSM.
It requires Xcalibur and Microsoft .NET 2.0 or later to be installed.
It is not clear whether it can be made to run stand-alone, on a system without a full installation of Xcalibur.
The unique feature of Raw2MSM is
that it improves the precursor mass accuracy by intensity-weighting the measured masses over their LC
elution profile and correcting with a lock mass. The approach is described in
Olsen, J. V.,
et al., Parts per million mass accuracy on an orbitrap mass spectrometer via lock mass injection into a
C-trap, Mol. & Cell. Proteomics 4 2010-2021 (2005).
Acknowledgements
Sequest is a registered trademark of the University of Washington.
Xcalibur is a registered trademark and Bioworks and Proteome Discoverer are trademarks of Thermo Electron Corporation.
|